Fluorescence in situ hybridization (FISH) is a cytogenetic technique used to detect and localize the presence or absence of specific DNA sequences on chromosomes. FISH uses fluorescent probes, each tagged with a different fluorophore, that bind to specific parts of the chromosome. Through multi-band fluorescence microscopy the positions where the fluorescent probes bound to the chromosomes can be displayed so as to derive information of clinical relevance based on the presence and position of the fluorescent probes.
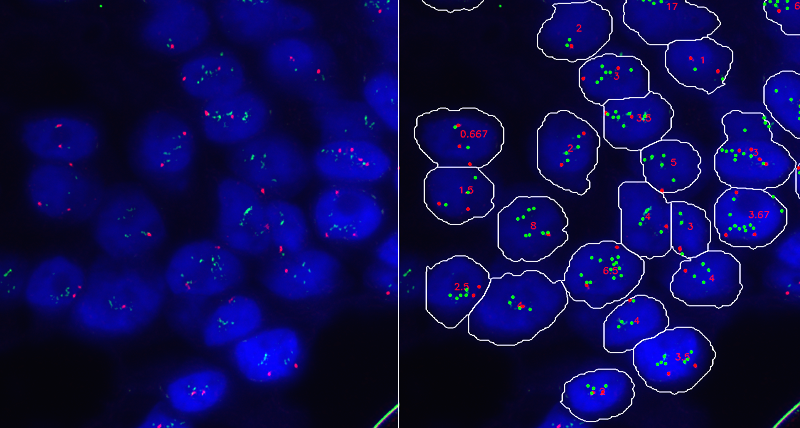
Accurate Evaluation of HER-2 Amplification in FISH Images
A sample application of this technique targets the measurement of the amplification of the HER-2 gene within the chromosomes, that constitutes a valuable indicator of invasive breast carcinomas. This requires the application to a tumor tissue sample of fluorescent probes that attach themselves to the HER-2 genes in a process called hybridization. These fluorescent probes carry a marker that emit light when the probes bind to the HER-2 genes, and this makes them visible as green spots under a fluorescent microscope. Similarly, a different probe, carrying a marker that makes it visible as a orange spot under a fluorescent microscope, is used to target the centromere 17 (CEP-17). Measuring the ratio of HER-2 over CEP-17 dots within each nucleus and then averaging this ratio for a representative number of cells allows estimation of HER-2 amplification.
In this research we present a system that supports accurate estimation of the ratio of HER-2 over CEP-17 dots in FISH images of breast tissue samples. Compared to previous work, the system incorporates a model to associate with each segmented nucleus a reliability score that estimates the confidence of the measure of the ratio of HER-2 over CEP-17 dots within the nucleus. This enables the computation of values of the ratio using only nuclei with high reliability scores so as to extract a measure of the amplification of HER-2 versus CEP-17 dots that better conforms to the evaluation of the pathologist compared to the ratio averaged over all available nuclei.